A comparative study of aptamer isolation by conventional and microfluidic strategies†
Abstract
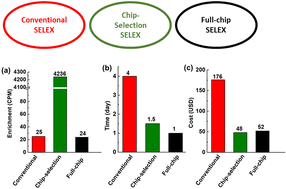
Aptamers are single-stranded oligonucleotide molecules that bind with high affinity and specificity to a wide range of target molecules. The method of systematic evolution of ligands by exponential enrichment (SELEX) plays an essential role in the isolation of aptamers from a randomized oligonucleotide library. To date, significant modifications and improvements of the SELEX process have been achieved, engendering various forms of SELEX from conventional SELEX to microfluidics-based full-chip SELEX. While full-chip SELEX is generally considered advantageous over conventional SELEX, there has not yet been a conclusive comparison between the methods. Herein, we present a comparative study of three SELEX strategies for aptamer isolation, including those using conventional agarose bead-based partitioning, microfluidic affinity selection, and fully integrated microfluidic affinity selection and PCR amplification. Using immunoglobulin E (IgE) as a model target molecule, we compare these strategies in terms of the time and cost for each step of the SELEX process including affinity selection, amplification, and oligonucleotide conditioning. Target-binding oligonucleotides in the enriched pools are sequenced and compared to assess the relative efficacy of the SELEX strategies. We show that the microfluidic strategies are more time- and cost-efficient than conventional SELEX.
- This article is part of the themed collection: Analyst HOT Articles 2023


 Please wait while we load your content...
Please wait while we load your content...