How proton transfer impacts hachimoji DNA†
Abstract
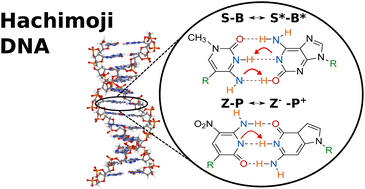
Hachimoji DNA is a synthetic nucleic acid extension of DNA, formed by an additional four bases, Z, P, S, and B, that can encode information and sustain Darwinian evolution. In this paper, we aim to look into the properties of hachimoji DNA and investigate the probability of proton transfer between the bases, resulting in base mismatch under replication. First, we present a proton transfer mechanism for hachimoji DNA, analogous to the one presented by Löwdin years prior. Then, we use density functional theory to calculate proton transfer rates, tunnelling factors and the kinetic isotope effect in hachimoji DNA. We determined that the reaction barriers are sufficiently low that proton transfer is likely to occur even at biological temperatures. Furthermore, the rates of proton transfer of hachimoji DNA are much faster than in Watson–Crick DNA due to the barrier for Z–P and S–B being 30% lower than in G–C and A–T. Suggesting that proton transfer occurs more frequently in hachimoji DNA than canonical DNA, potentially leading to a higher mutation rate.


 Please wait while we load your content...
Please wait while we load your content...