Novel fluorescence-based method for rapid quantification of live bacteria in river water and treated wastewater†
Abstract
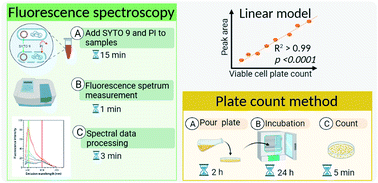
Monitoring bacteria is essential for ensuring microbial safety of water sources, including river water and treated wastewater. The plate count method is common for monitoring bacterial abundance, although it cannot detect all live bacteria such as viable but non-culturable bacteria, causing underestimation of microbial risks. Live/Dead BacLight kit, involving fluorochromes SYTO 9 and propidium iodide (PI), provides an alternative to assess bacterial viability using flow cytometry or microscopy. However, its application is limited due to the high cost of flow cytometry and the inapplicability of microscopy to most environmental waters. Thus, this study introduces the combination of BacLight kit and fluorescence spectroscopy for quantifying live bacteria in river water and treated wastewater. Mixtures of live and dead Escherichia coli (E. coli) with various ratios and total cell concentrations were stained with SYTO 9 and PI and measured by fluorescence spectroscopy. The fluorescence emission peak area of SYTO 9 in the range of 500–510 nm at the excitation wavelength of 470 nm correlates linearly with the viable cell counts (R2 > 0.99, p < 0.0001) with only slight variations in the complex water matrix. The tested method can quantify the live E. coli from 3.67 × 104 to 2.70 × 107 cells per mL. This method is simple, sensitive and reliable for quantifying live bacteria in environmental water, which can be later integrated into real-time monitoring systems.
- This article is part of the themed collection: Topic Collection: Sensors, Detection and Monitoring


 Please wait while we load your content...
Please wait while we load your content...