Bisulfite-free and single-nucleotide resolution sequencing of DNA epigenetic modification of 5-hydroxymethylcytosine using engineered deaminase†
Abstract
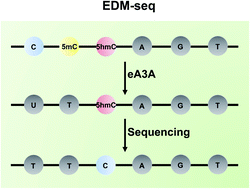
The discovery of 5-hydroxymethylcytosine (5hmC) in mammalian genomes is a landmark in epigenomics study. Similar to 5-methylcytosine (5mC), 5hmC is viewed as a critical epigenetic modification. Deciphering the functions of 5hmC necessitates the location analysis of 5hmC in genomes. Here, we proposed an engineered deaminase-mediated sequencing (EDM-seq) method for the quantitative detection of 5hmC in DNA at single-nucleotide resolution. This method capitalizes on the engineered human apolipoprotein B mRNA-editing catalytic polypeptide-like 3A (A3A) protein to produce differential deamination activity toward cytosine, 5mC, and 5hmC. In EDM-seq, the engineered A3A (eA3A) protein can deaminate C and 5mC but not 5hmC. The original C and 5mC in DNA are deaminated by eA3A to form U and T, both of which are read as T during sequencing, while 5hmC is resistant to deamination by eA3A and is still read as C during sequencing. Therefore, the remaining C in the sequence manifests the original 5hmC. By EDM-seq, we achieved the quantitative detection of 5hmC in genomic DNA of lung cancer tissue. The EDM-seq method is bisulfite-free and does not require DNA glycosylation or chemical treatment, which offers a valuable tool for the straightforward and quantitative detection of 5hmC in DNA at single-nucleotide resolution.


 Please wait while we load your content...
Please wait while we load your content...