DNA origami book biosensor for multiplex detection of cancer-associated nucleic acids†
Abstract
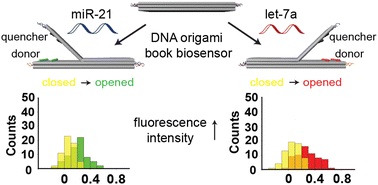
DNA nanotechnology provides a promising approach for the development of biomedical point-of-care diagnostic nanoscale devices that are easy to use and cost-effective, highly sensitive and thus constitute an alternative to expensive, complex diagnostic devices. Moreover, DNA nanotechnology-based devices are particularly advantageous for applications in oncology, owing to being ideally suited for the detection of cancer-associated nucleic acids, including circulating tumor-derived DNA fragments (ctDNAs), circulating microRNAs (miRNAs) and other RNA species. Here, we present a dynamic DNA origami book biosensor that is precisely decorated with arrays of fluorophores acting as donors and acceptors and also fluorescence quenchers that produce a strong optical readout upon exposure to external stimuli for the single or dual detection of target oligonucleotides and miRNAs. This biosensor allowed the detection of target molecules either through the decrease of Förster resonance energy transfer (FRET) or an increase in the fluorescence intensity profile owing to a rotation of the constituent top layer of the structure. Single-DNA origami experiments showed that detection of two targets can be achieved simultaneously within 10 min with a limit of detection in the range of 1–10 pM. Overall, our DNA origami book biosensor design showed sensitive and specific detection of synthetic target oligonucleotides and natural miRNAs extracted from cancer cells. Based on these results, we foresee that our DNA origami biosensor may be developed into a cost-effective point-of-care diagnostic strategy for the specific and sensitive detection of a variety of DNAs and RNAs, such as ctDNAs, miRNAs, mRNAs, and viral DNA/RNAs in human samples.


 Please wait while we load your content...
Please wait while we load your content...