Activating the p53 anti-cancer pathway by targeting the MDM2/MDMX dimer interface with short peptide segments: a computational peptide design experiment
Abstract
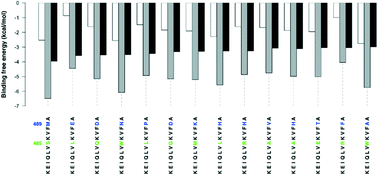
Formation of the MDM2/MDMX heterodimer through association of the RING domains of the MDM2 and MDMX proteins is essential for p53 activation. Disrupting the formation of this heterodimer is, therefore, a promising approach for p53 reactivation therapy. Here, we use blind docking simulations of dodecapeptide segments to the MDM2 RING domain, in order to assess their potential to disrupt the MDM2/MDMX formation. We used the KEIQLVIKVFI489A and KEIQLVIKVAE489A dodecapeptides as reference and control peptides since the former is reported to activate the p53 pathway through reducing p53 ubiquitination, while the latter has no effect on p53 ubiquitination. We could rationalize the difference between the activities of the two peptides in terms of preferential binding (ΔΔG) to the MDM2/MDMX interface hotspot residues compared to binding to sites outside the interface region. Binding of the KEIQLVIKVFI489A reference peptide to the MDM2/MDMX interface region was found to be more stable, ΔG = −3.95 kcal mol−1, than its binding to the outside of the interface, ΔG= −2.93 kcal mol−1, indicating preferential binding (ΔΔG = −1.02 kcal mol−1) of the KEIQLVIKVFI489A reference peptide to the MDM2/MDMX interface. By contrast, binding of the KEIQLVIKVAE489A control peptide to the MDM2/MDMX interface region is less stable, ΔG = −1.50 kcal mol−1, than its binding to the outside of the interface, ΔG = −2.29 kcal mol−1, indicating preferential binding to sites outside the interface region (ΔΔG = +0.79 kcal mol−1). Preferential binding (ΔΔG) could, therefore, be a better metric for the design and identification of new peptide leads. Systematic mutation of the I485 and I489 residues of the reference peptide leads to identification of 14 mutant peptides that show at least three-fold preferential binding to the MDM2/MDMX interface (ΔΔG ∼ −3.00 kcal mol−1) lower than the KEIQLVIKVFI489A reference peptide (ΔΔG = −1.02 kcal mol−1), with the KEIQLVSKVFI489M mutant being the most stable (ΔΔG = −3.96 kcal mol−1). These peptides could, therefore, provide lead structures for p53 reactivation therapy.


 Please wait while we load your content...
Please wait while we load your content...