Automated biophysical classification of apoptotic pancreatic cancer cell subpopulations by using machine learning approaches with impedance cytometry†
Abstract
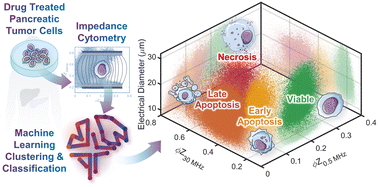
Unrestricted cell death can lead to an immunosuppressive tumor microenvironment, with dysregulated apoptotic signaling that causes resistance of pancreatic cancer cells to cytotoxic therapies. Hence, modulating cell death by distinguishing the progression of subpopulations under drug treatment from viable towards early apoptotic, late apoptotic, and necrotic states is of interest. While flow cytometry after fluorescent staining can monitor apoptosis with single-cell sensitivity, the background of non-viable cells within non-immortalized pancreatic tumors from xenografts can confound distinction of the intensity of each apoptotic state. Based on single-cell impedance cytometry of drug-treated pancreatic cancer cells that are obtained from tumor xenografts with differing levels of gemcitabine sensitivity, we identify the biophysical metrics that can distinguish and quantify cellular subpopulations at the early apoptotic versus late apoptotic and necrotic states, by using machine learning methods to train for the recognition of each phenotype. While supervised learning has previously been used for classification of datasets with known classes, our advancement is the utilization of optimal positive controls for each class, so that clustering by unsupervised learning and classification by supervised learning can occur on unknown datasets, without human interference or manual gating. In this manner, automated biophysical classification can be used to follow the progression of apoptotic states in each heterogeneous drug-treated sample, for developing drug treatments to modulate cancer cell death and advance longitudinal analysis to discern the emergence of drug resistant phenotypes.
- This article is part of the themed collections: Machine Learning and Artificial Intelligence: A cross-journal collection and AI in Microfluidics


 Please wait while we load your content...
Please wait while we load your content...