Towards rational nanomaterial design by predicting drug–nanoparticle system interaction vs. bacterial metabolic networks†
Abstract
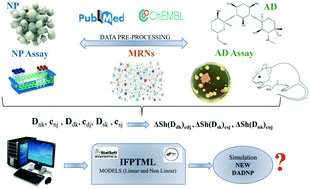
The emergence of multidrug-resistant (MDR) strains with perturbed metabolic networks (MNs) pushes researchers to improve antibacterial drugs (ADs). Certain nanoparticles (NPs) may present antibacterial activity along with acting as delivery systems. Thus, developing dual antibacterial drug–nanoparticle (DADNP) systems becomes an option. However, testing DADNPs vs. strains with different MNs is a hard and costly task. Artificial intelligence (AI) or machine learning (ML) could accelerate this by predicting bacterial sensitivity. In this work, we used an information fusion perturbation-theory machine learning (IFPTML) analysis and mapping of DADNP (AD + NP) systems vs. MNs of pathogenic bacterial species as a new application of AI/ML methods. Furthermore, most existing AI/ML models do not use cj of experimental conditions of assays (i.e., bacteria species, strain, NP shape, etc.) as input vectors. A working solution may be the use of an AI/ML method with an information fusion (IF) additive approach. Additive IF uses the sets of vectors Ddk, Dnk, Dmk and cdk, cnk, csk as inputs with information about AD, NP, and MN structure and assays separately. Accordingly, the IFPTML algorithm was selected to seek predictive models based on a ChEMBL dataset of >160 000 AD assays enriched with 300 NP assays and >25 MNs of different bacterial species. IFPTML uses the IF process to join the three datasets, PT operators (PTOs) to codify Ddk, Dnk, Dsk and cdk, cnk, csk vector information, and ML algorithms to train the model. The IFPTML linear discriminant analysis (LDA) model with Sp ≈ 90% and Sn ≈ 80% and the best artificial neural network (ANN) model found with Sp ≈ Sn ≈ 95% in the training/validation series presented good results. This kind of model could be useful for DADNP system discovery. We also ran a simulation with >140 000 points of putative DADNP systems vs. wild type and knockout (KO) computationally generated bacterial strains. The linear and additive IFPTML model was able to predict 102 experimental cases of complex DADNPs with a high degree of structural and biological variety. This led us to introduce the concept of MDR computational surveillance that could help to detect new strains of MDR bacteria.


 Please wait while we load your content...
Please wait while we load your content...