A fast–slow method to treat solute dynamics in explicit solvent†
Abstract
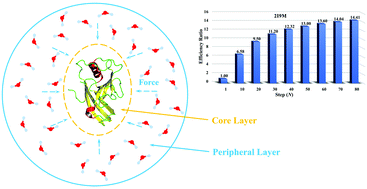
Aiming to reduce the computational cost in the current explicit solvent molecular dynamics (MD) simulation, this paper proposes a fast–slow method for the fast MD simulation of biomolecules in explicit solvent. This fast–slow method divides the entire system into two parts: a core layer (typically solute or biomolecule) and a peripheral layer (typically solvent molecules). The core layer is treated using standard MD method but the peripheral layer is treated by a slower dynamics method to reduce the computational cost. We compared four different simulation models in testing calculations for several small proteins. These include gas-phase, implicit solvent, fast–slow explicit solvent and standard explicit solvent MD simulations. Our study shows that gas-phase and implicit solvent models do not provide a realistic solvent environment and fail to correctly produce reliable dynamic structures of proteins. On the other hand, the fast–slow method can essentially reproduce the same solvent effect as the standard explicit solvent model while gaining an order of magnitude in efficiency. This fast–slow method thus provides an efficient approach for accelerating the MD simulation of biomolecules in explicit solvent.


 Please wait while we load your content...
Please wait while we load your content...