Cryo-EM structure of acylpeptide hydrolase reveals substrate selection by multimerization and a multi-state serine-protease triad†
Abstract
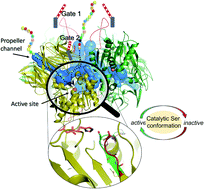
The first structure of tetrameric mammalian acylaminoacyl peptidase, an enzyme that functions as an upstream regulator of the proteasome through the removal of terminal N-acetylated residues from its protein substrates, was determined by cryo-EM and further elucidated by MD simulations. Self-association results in a toroid-shaped quaternary structure, guided by an amyloidogenic β-edge and unique inserts. With a Pro introduced into its central β-sheet, sufficient conformational freedom is awarded to the segment containing the catalytic Ser587 that the serine protease catalytic triad alternates between active and latent states. Active site flexibility suggests that the dual function of catalysis and substrate selection are fulfilled by a novel mechanism: substrate entrance is regulated by flexible loops creating a double-gated channel system, while binding of the substrate to the active site is required for stabilization of the catalytic apparatus – as a second filter before hydrolysis. The structure not only underlines that within the family of S9 proteases homo-multimerization acts as a crucial tool for substrate selection, but it will also allow drug design targeting of the ubiquitin-proteasome system.


 Please wait while we load your content...
Please wait while we load your content...