Distinct core glycan and O-glycoform utilization of SARS-CoV-2 Omicron variant Spike protein RBD revealed by top-down mass spectrometry†
Abstract
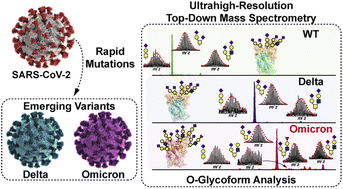
The SARS-CoV-2 Omicron (B.1.1.529) variant possesses numerous spike (S) mutations particularly in the S receptor-binding domain (S-RBD) that significantly improve transmissibility and evasion of neutralizing antibodies. But exactly how the mutations in the Omicron variant enhance viral escape from immunological protection remains to be understood. The S-RBD remains the principal target for neutralizing antibodies and therapeutics, thus new structural insights into the Omicron S-RBD and characterization of the post-translational glycosylation changes can inform rational design of vaccines and therapeutics. Here we report the molecular variations and O-glycoform changes of the Omicron S-RBD variant as compared to wild-type (WA1/2020) and Delta (B.1.617.2) variants using high-resolution top-down mass spectrometry (MS). A novel O-glycosite (Thr376) unique to the Omicron variant is identified. Moreover, we have directly quantified the Core 1 and Core 2 O-glycan structures and characterized the O-glycoform structural heterogeneity of the three variants. Our findings reveal high resolution detail of Omicron O-glycoforms and their utilization to provide direct molecular evidence of proteoform alterations in the Omicron variant which could shed light on how this variant escapes immunological protection.


 Please wait while we load your content...
Please wait while we load your content...