Stain-free Gram staining classification of pathogens via single-cell Raman spectroscopy combined with machine learning†
Abstract
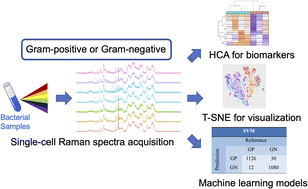
Gram staining (GS) is one of the routine microbiological operations to classify bacteria based on the cell wall structure. Accurate GS classification of pathogens is of great significance since it helps correct administration of antimicrobial treatment. The laborious procedure and low sensitivity results related to conventional GS have resulted in reluctance among clinicians. In this study, we integrate confocal Raman spectroscopy and machine learning techniques to distinguish Gram-negative (GN) or Gram-positive (GP) bacteria. A single-cell Raman database including seven most common clinical pathogens (three GP strains and four GN strains) was constructed. Machine learning algorithms including the support-vector machine (SVM), k-nearest neighbors' algorithm (k-NN), gradient boosting machine (GBM), linear discriminant analysis (LDA), and t-distributed stochastic neighbor embedding (t-SNE) were trained to achieve the binary classification for GS. With such a relatively small database, the SVM model achieved the highest accuracy of 98.1%. The molecular signatures of GN and GP embedded in their Raman fingerprints were identified with hierarchical cluster analysis (HCA). The results indicated that Raman peaks for peptidoglycan and teichoic acid were the most significant factors that contributed to accurate classification. The Raman machine learning approach could greatly enhance the diagnosis of pathogenic infections.


 Please wait while we load your content...
Please wait while we load your content...