Genomics-driven discovery of chiral triscatechol siderophores with enantiomeric Fe(iii) coordination†
Abstract
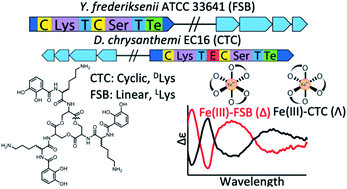
Ferric complexes of triscatechol siderophores may assume one of two enantiomeric configurations at the iron site. Chirality is known to be important in the iron uptake process, however an understanding of the molecular features directing stereospecific coordination remains ambiguous. Synthesis of the full suite of (DHBL/DLysL/DSer)3 macrolactone diastereomers, which includes the siderophore cyclic trichrysobactin (CTC), enables the effects that the chirality of Lys and Ser residues exert on the configuration of the Fe(III) complex to be defined. Computationally optimized geometries indicate that the Λ/Δ configurational preferences are set by steric interactions between the Lys sidechains and the peptide backbone. The ability of each (DHBL/DLysL/DSer)3 diastereomer to form a stable Fe(III) complex prompted a genomic search for biosynthetic gene clusters (BGCs) encoding the synthesis of these diastereomers in microbes. The genome of the plant pathogen Dickeya chrysanthemi EC16 was sequenced and the genes responsible for the biosynthesis of CTC were identified. A related but distinct BGC was identified in the genome of the opportunistic pathogen Yersinia frederiksenii ATCC 33641; isolation of the siderophore from Y. frederiksenii ATCC 33641, named frederiksenibactin (FSB), revealed the triscatechol oligoester, linear-(DHBLLysLSer)3. Circular dichroism (CD) spectroscopy establishes that Fe(III)–CTC and Fe(III)–FSB are formed in opposite enantiomeric configuration, consistent with the results of the ferric complexes of the cyclic (DHBL/DLysL/DSer)3 diastereomers.


 Please wait while we load your content...
Please wait while we load your content...