Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology
Abstract
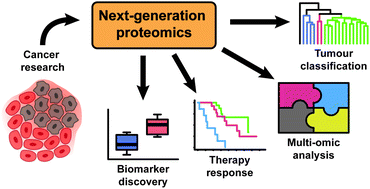
Data-independent acquisition mass spectrometry (DIA-MS) is a next generation proteomic methodology that generates permanent digital proteome maps offering highly reproducible retrospective analysis of cellular and tissue specimens. The adoption of this technology has ushered a new wave of oncology studies across a wide range of applications including its use in molecular classification, oncogenic pathway analysis, drug and biomarker discovery and unravelling mechanisms of therapy response and resistance. In this review, we provide an overview of the experimental workflows commonly used in DIA-MS, including its current strengths and limitations versus conventional data-dependent acquisition mass spectrometry (DDA-MS). We further summarise a number of key studies to illustrate the power of this technology when applied to different facets of oncology. Finally we offer a perspective of the latest innovations in DIA-MS technology and machine learning–based algorithms necessary for driving the development of high-throughput, in-depth and reproducible proteomic assays that are compatible with clinical diagnostic workflows, which will ultimately enable the delivery of precision cancer medicine to achieve optimal patient outcomes.


 Please wait while we load your content...
Please wait while we load your content...