An interactive Python-based data processing platform for single particle and single cell ICP-MS†
Abstract
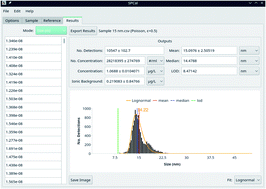
Single particle (SP) and single cell (SC) inductively coupled plasma-mass spectrometry (ICP-MS) are gaining increasing momentum in environmental and medical sciences for the analysis of nanoparticles, microstructures, and individual cells. This work presents an open-source Python-based SP/SC ICP-MS data processing platform with an interactive graphical user interface. The program guides users through the analysis of large data sets and uses efficient and transparent algorithms. Gaussian and Poisson-based data filtering enables fit for purpose thresholding of particle signals from background noise. Implementation of windowed filters extends applicability of the software to SP laser ablation-ICP-MS and other data sets that contain drifting or variable backgrounds. SP or SC signals recorded with multiple data points are integrated and several distinct calibration and processing pathways may be used to determine masses, sizes, and number concentrations, or to calculate intracellular concentrations. Relevant parameters including means, medians, ionic background levels and limits of analysis are automatically calculated and visualised together with histograms of raw and calibrated data. As a proof of principle, the developed data processing platform was employed to characterise TiO2 nanoparticles in surface water, microplastic particles in soil and the C content across individual cells.


 Please wait while we load your content...
Please wait while we load your content...