Computational modeling of dynein motor proteins at work
Abstract
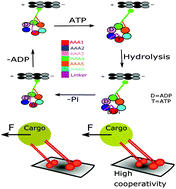
Along with various experimental methods, a combination of theoretical and computational methods is essential to explore different length-scale and time-scale processes in the biological system. The functional mechanism of a dynein, an ATP-fueled motor protein, working in a multiprotein complex, involves a wide range of length/time-scale events. It generates mechanical force from chemical energy and moves on microtubules towards the minus end direction while performing a large number of biological processes including ciliary beating, intracellular material transport, and cell division. Like in the cases of other conventional motor proteins, a combination of experimental techniques including X-crystallography, cryo-electron microscopy, and single molecular assay have provided a wealth of information about the mechanochemical cycle of a dynein. Dyneins have a large and complex structural architecture and therefore, computational modeling of different aspects of a dynein is extremely challenging. As the process of dynein movement involves varying length and timescales, it demands, like in experiments, a combination of computational methods covering such a wide range of processes for the comprehensive investigation of the mechanochemical cycle. In this review article, we will summarize how the use of state-of-the-art computational methods can provide a detailed molecular understanding of the mechanochemical cycle of the dynein. We implemented all-atom molecular dynamics simulations and hybrid quantum-mechanics/molecular-mechanics simulations to explore the ATP hydrolysis mechanisms at the primary ATPase site (AAA1) of dynein. To investigate the large-scale conformational changes we employed coarse-grained structure-based molecular dynamics simulations to capture the domain motions. Here we explored the conformational changes upon binding of ATP at AAA1, nucleotide state-dependent regulation of the mechanochemical cycle, and inter-head coordination by inter-head tension. Additionally, implementing a phenomenological theoretical model we explore the force-dependent detachment rate of a motorhead from the microtubule and the principle of multi-dynein cooperation during cargo transport.
- This article is part of the themed collection: 2021 Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...