Bioorthogonal protein labelling enables the study of antigen processing of citrullinated and carbamylated auto-antigens†
Abstract
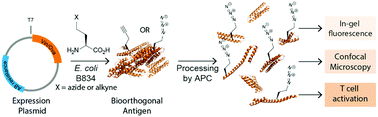
Proteolysis is fundamental to many biological processes. In the immune system, it underpins the activation of the adaptive immune response: degradation of antigenic material into short peptides and presentation thereof on major histocompatibility complexes, leads to activation of T-cells. This initiates the adaptive immune response against many pathogens. Studying proteolysis is difficult, as the oft-used polypeptide reporters are susceptible to proteolytic sequestration themselves. Here we present a new approach that allows the imaging of antigen proteolysis throughout the processing pathway in an unbiased manner. By incorporating bioorthogonal functionalities into the protein in place of methionines, antigens can be followed during degradation, whilst leaving reactive sidechains open to templated and non-templated post-translational modifications, such as citrullination and carbamylation. Using this approach, we followed and imaged the post-uptake fate of the commonly used antigen ovalbumin, as well as the post-translationally citrullinated and/or carbamylated auto-antigen vinculin in rheumatoid arthritis, revealing differences in antigen processing and presentation.
- This article is part of the themed collection: RSC Chemical Biology Transparent Peer Review Collection


 Please wait while we load your content...
Please wait while we load your content...