TMT-based quantitative proteome profiles reveal the memory function of a whole heart decellularized matrix for neural stem cell trans-differentiation into the cardiac lineage†
Abstract
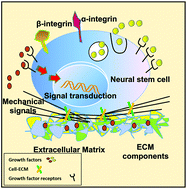
Whole organ or tissue decellularized matrices are a promising scaffold for tissue engineering because they maintain the specific memory of the original organ or tissue. A whole organ or tissue decellularized matrix contains extracellular matrix (ECM) components, and exhibits ultrastructural and mechanical properties, which could significantly regulate the fate of stem cells. To better understand the memory function of whole organ decellularized matrices, we constructed a heart decellularized matrix and seeded cross-embryonic layer stem cells – neural stem cells (NSCs) to repopulate the matrix, engineering cardiac tissue, in which a large number of NSCs differentiated into the neural lineage, but besides that, NSCs showed an obvious tendency of trans-differentiating into cardiac lineage cells. The results demonstrated that the whole heart decellularized microenvironment possesses memory function. To reveal the underlying mechanism, TMT-based quantitative proteomics analysis was used to identify the differently expressed proteins in the whole heart decellularized matrix compared with a brain decellularized matrix. 937 of the proteins changed over 1.5 fold, with 573 of the proteins downregulated and 374 of the proteins upregulated, among which integrin ligands in the ECM serve as key signals in regulating NSC fate. The findings here provide a novel insight into the memory function of tissue-specific microenvironments and pave the way for the therapeutic application of personalized tissues.


 Please wait while we load your content...
Please wait while we load your content...