Differences in SMA-like polymer architecture dictate the conformational changes exhibited by the membrane protein rhodopsin encapsulated in lipid nano-particles†
Abstract
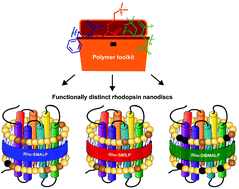
Membrane proteins are of fundamental importance to cellular processes and nano-encapsulation strategies that preserve their native lipid bilayer environment are particularly attractive for studying and exploiting these proteins. Poly(styrene-co-maleic acid) (SMA) and related polymers poly(styrene-co-(N-(3-N′,N′-dimethylaminopropyl)maleimide)) (SMI) and poly(diisobutylene-alt-maleic acid) (DIBMA) have revolutionised the study of membrane proteins by spontaneously solubilising membrane proteins direct from cell membranes within nanoscale discs of native bilayer called SMA lipid particles (SMALPs), SMILPs and DIBMALPs respectively. This systematic study shows for the first time, that conformational changes of the encapsulated protein are dictated by the solubilising polymer. The photoactivation pathway of rhodopsin (Rho), a G-protein-coupled receptor (GPCR), comprises structurally-defined intermediates with characteristic absorbance spectra that revealed conformational restrictions with styrene-containing SMA and SMI, so that photoactivation proceeded only as far as metarhodopsin-I, absorbing at 478 nm, in a SMALP or SMILP. In contrast, full attainment of metarhodopsin-II, absorbing at 382 nm, was observed in a DIBMALP. Consequently, different intermediate states of Rho could be generated readily by simply employing different SMA-like polymers. Dynamic light-scattering and analytical ultracentrifugation revealed differences in size and thermostability between SMALP, SMILP and DIBMALP. Moreover, encapsulated Rho exhibited different stability in a SMALP, SMILP or DIBMALP. Overall, we establish that SMA, SMI and DIBMA constitute a ‘toolkit’ of solubilising polymers, so that selection of the appropriate solubilising polymer provides a spectrum of useful attributes for studying membrane proteins.


 Please wait while we load your content...
Please wait while we load your content...