Key residues of the receptor binding domain in the spike protein of SARS-CoV-2 mediating the interactions with ACE2: a molecular dynamics study
Abstract
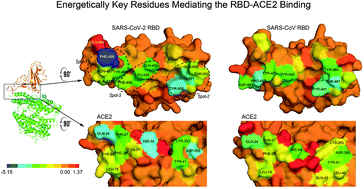
The widespread coronavirus disease 2019 (COVID-19) has been declared a global health emergency. As one of the most important targets for antibody and drug developments, the Spike RBD-ACE2 interface has received extensive attention. Here, using molecular dynamics simulations, we explicitly analyzed the energetic features of the RBD-ACE2 complex of both SARS-CoV and SARS-CoV-2. Despite the high structural similarity, the binding strength of SARS-CoV-2 to the ACE2 receptor is estimated to be −16.35 kcal mol−1 stronger than that of SARS-CoV. Energy decomposition analyses identified three binding patches in SARS-CoV-2 RBD and eleven key residues (F486, Y505, N501, Y489, Q493, L455, etc.), which are believed to be the main targets for drug development. The dominating forces arise from van der Waals attractions and dehydration of these residues. Compared with SARS-CoV, we found seven mutational sites (K417, L455, A475, G476, E484, Q498 and V503) on SARS-CoV-2 that unexpectedly weakened the RBD-ACE2 binding. Interestingly, the E484 site is recognized to be the most repulsive residue at the RBD-ACE2 interface, indicating that from the energy point of view, a mutation of E484 would be beneficial to RBD-ACE2 binding. This is in line with recent findings that it is mutated by lysine (E484K mutation) in the rapidly spreading variants of COVID-19 belonging to the B.1.351 and P.1 lineages. In addition, this mutation is reported to cause virus neutralization escapes from highly neutralizing COVID-19 convalescent plasma. Thus, further efforts are required to probe its functional relevance. Overall, our results present a systematic understanding of the energetic binding features of SARS-CoV-2 RBD with the ACE2 receptor, which can provide a valuable insight for the design of SARS-CoV-2 drugs and identification of cross-active antibodies.


 Please wait while we load your content...
Please wait while we load your content...