Systematic investigation of mouse models of Parkinson's disease by transcriptome mapping on a brain-specific genome-scale metabolic network†
Abstract
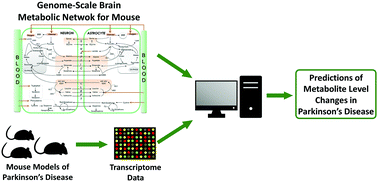
Genome-scale metabolic networks enable systemic investigation of metabolic alterations caused by diseases by providing interpretation of omics data. Although Mus musculus (mouse) is one of the most commonly used model organisms for neurodegenerative diseases, a brain-specific metabolic network model of mice has not yet been reconstructed. Here we reconstructed the first brain-specific metabolic network model of mice, iBrain674-Mm, by a homology-based approach, which consisted of 992 reactions controlled by 674 genes and distributed over 48 pathways. We validated the newly reconstructed network model by showing that it predicts healthy resting-state metabolic phenotypes of mouse brain compatible with the literature. We later used iBrain674-Mm to interpret various experimental mouse models of Parkinson's Disease (PD) at the transcriptome level. To this end, we applied a constraint-based modelling based biomarker prediction method called TIMBR (Transcriptionally Inferred Metabolic Biomarker Response) to predict altered metabolite production from transcriptomic data. Systemic analysis of seven different PD mouse models by TIMBR showed that the neuronal levels of glutamate, lactate, creatine phosphate, neuronal acetylcholine, bilirubin and formate increased in most of the PD mouse models, whereas the levels of melatonin, epinephrine, astrocytic formate and astrocytic bilirubin decreased. Although most of the predictions were consistent with the literature, there were some inconsistencies among different PD mouse models, signifying that there is no perfect experimental model to reflect PD metabolism. The newly reconstructed brain-specific genome-scale metabolic network model of mice can make important contributions to the interpretation and development of experimental mouse models of PD and other neurodegenerative diseases.


 Please wait while we load your content...
Please wait while we load your content...