Incorporation efficiency and inhibition mechanism of 2′-substituted nucleotide analogs against SARS-CoV-2 RNA-dependent RNA polymerase†
Abstract
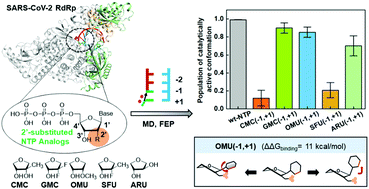
The ongoing pandemic caused by SARS-CoV-2 emphasizes the need for effective therapeutics. Inhibition of SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) by nucleotide analogs provides a promising antiviral strategy. One common group of RdRp inhibitors, 2′-modified nucleotides, are reported to exhibit different behaviors in the SARS-CoV-2 RdRp transcription assay. Three of these analogs, 2′-O-methyl UTP, Sofosbuvir, and 2′-methyl CTP, act as effective inhibitors in previous biochemical experiments, while Gemcitabine and ara-UTP show no inhibitory activity. To understand the impact of the 2′-modification on their inhibitory effects, we conducted extensive molecular dynamics simulations and relative binding free energy calculations using the free energy perturbation method on SARS-CoV-2 replication-transcription complex (RTC) with these five nucleotide analogs. Our results reveal that the five nucleotide analogs display comparable binding affinities to SARS-CoV-2 RdRp and they can all be added to the nascent RNA chain. Moreover, we examine how the incorporation of these nucleotide triphosphate (NTP) analogs will impact the addition of the next nucleotide. Our results indicate that 2′-O-methyl UTP can weaken the binding of the subsequent NTP and consequently lead to partial chain termination. Additionally, Sofosbuvir and 2′-methyl CTP can cause immediate termination due to the strong steric hindrance introduced by their bulky 2′-methyl groups. In contrast, nucleotide analogs with smaller substitutions, such as the fluorine atoms and the ara-hydroxyl group in Gemcitabine and ara-UTP, have a marginal impact on the polymerization process. Our findings are consistent with experimental observations, and more importantly, shed light on the detailed molecular mechanism of SARS-CoV-2 RdRp inhibition by 2′-substituted nucleotide analogs, and may facilitate the rational design of antiviral agents to inhibit SARS-CoV-2 RdRp.
- This article is part of the themed collection: 2021 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...