A novel artificial intelligence protocol for finding potential inhibitors of acute myeloid leukemia†
Abstract
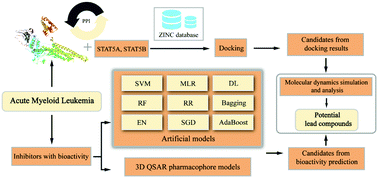
There is currently no effective treatment for acute myeloid leukemia, and surgery is also ineffective as an important treatment for most tumors. Rapidly developing artificial intelligence technology can be applied to different aspects of drug development, and it plays a key role in drug discovery. Based on network pharmacology and virtual screening, candidates were selected from the molecular database. Nine artificial intelligence algorithm models were used to further verify the candidates’ potential. The 350 training results of the deep learning model showed higher credibility, and the R-square of the training set and test set of the optimal model reached 0.89 and 0.84, respectively. The random forest model has an R-square of 0.91 and a mean square error of only 0.003. The R-square of the Adaptive Boosting model and the Bagging model reached 0.92 and 0.88, respectively. Molecular dynamics simulation evaluated the stability of the ligand–protein complex and achieved good results. Artificial intelligence models had unearthed the promising candidates for STAT3 inhibitors, and the good performance of most models showed that they still had practical value on small data sets.


 Please wait while we load your content...
Please wait while we load your content...