Proteomics and metabolomics analysis reveal potential mechanism of extended-spectrum β-lactamase production in Escherichia coli†
Abstract
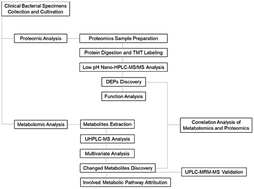
In this study, ten clinical susceptible strains and ten clinical ESBL-EC (extended-spectrum β-lactamase-producing Escherichia coli) were screened and obtained by microbial identification using ITEK® 2 Compact. TMT (Tandem Mass Tag) proteomics analysis discovered 1553 DEPs (differentially expressed proteins) between ESBL-EC and non-ESBL-EC. In addition, an untargeted metabolomics assay by using UHPLC-MS (ultra-high-performance liquid chromatography-mass spectrometry) was applied to compare the differential profiles of metabolites between β-lactam antibiotic-sensitive E. coli and multidrug-resistant ESBL-producing E. coli strains. The PCA (principal component analysis) score plots and OPLS-DA (orthogonal projections to latent structures discriminant analysis) plots clearly discriminated ESBL-EC and non-ESBL-EC, and volcano analysis presented 606 and 459 altered metabolites between ESBL-EC vs. non-ESBL-EC in positive and negative ion modes, respectively. Interestingly, the bioinformatics analysis demonstrated that the purine metabolism pathway was enriched in ESBL-EC. These results suggest that the existence of extended-spectrum β-lactamase affects the metabolite and protein profiles of E. coli. The correlation analysis of metabolomics and proteomics data established a correlation between DEPs and differential metabolites in the purine metabolism pathway. Moreover, three metabolite candidates in the purine metabolism pathway were validated by the UPLC-MRM-MS (ultra-performance liquid chromatography multiple reaction monitoring mass spectrometry) method. Our data suggest that these DEPs and differential metabolites may play important roles in the antibiotic resistance of ESBL-EC. Our study can provide scientific data for the mechanism study of antibiotic resistance of ESBL-EC at the metabolite and protein levels and targeting modulators to these pathways may be effective for treatment of ESBL-EC strains.


 Please wait while we load your content...
Please wait while we load your content...