Head-to-head comparison of LNA, MPγPNA, INA and Invader probes targeting mixed-sequence double-stranded DNA†
Abstract
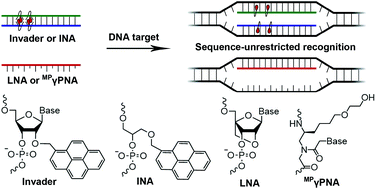
Four probe chemistries are characterized and compared with respect to thermal denaturation temperatures (Tms), thermodynamic parameters associated with duplex formation, and recognition of mixed-sequence double-stranded (ds) DNA targets: (i) oligodeoxyribonucleotides (ONs) modified with Locked Nucleic Acid (LNA) monomers, (ii) MPγPNAs, i.e., single-stranded peptide nucleic acid (PNA) probes that are functionalized at the γ-position with (R)-diethylene glycol (mini-PEG, MP) moieties, (iii) Invader probes, i.e., DNA duplexes modified with +1 interstrand zipper arrangements of 2′-O-(pyren-1-yl)methyl-RNA monomers, and (iv) intercalating nucleic acids (INAs), i.e., DNA duplexes with opposing insertions of 1-O-(1-pyrenylmethyl)glycerol bulges. Invader and INA probes, which are designed to violate the nearest-neighbor exclusion principle, denature readily, whereas the individual probe strands display exceptionally high affinity towards complementary DNA (cDNA) as indicated by increases in Tms of up to 8 °C per modification. Optimized Invader and INA probes enable efficient and highly specific recognition of mixed-sequence dsDNA targets with self-complementary regions (C50 = 30–50 nM), whereas recognition is less efficient with LNA-modified ONs and fully modified MPγPNAs due to lower cDNA affinity (LNA) and a proclivity for dimerization (LNA and MPγPNA). A Cy3-labeled Invader probe is shown to stain telomeric DNA of individual chromosomes in metaphasic spreads under non-denaturing conditions with excellent specificity.
- This article is part of the themed collection: Chemical Biology in OBC

 Please wait while we load your content...
Please wait while we load your content...