Making biological membrane resistant to the toxicity of misfolded protein oligomers: a lesson from trodusquemine†
Abstract
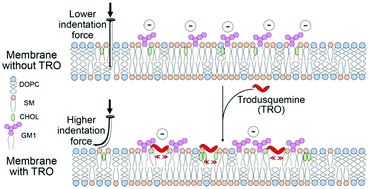
Trodusquemine is an aminosterol known to prevent the binding of misfolded protein oligomers to cell membranes and to reduce their toxicity in a wide range of neurodegenerative diseases. Its precise mechanism of action, however, remains unclear. To investigate this mechanism, we performed confocal microscopy, fluorescence resonance energy transfer (FRET) and nuclear magnetic resonance (NMR) measurements, which revealed a strong binding of trodusquemine to large unilamellar vesicles (LUVs) and neuroblastoma cell membranes. Then, by combining quartz crystal microbalance (QCM), fluorescence quenching and anisotropy, and molecular dynamics (MD) simulations, we found that trodusquemine localises within, and penetrates, the polar region of lipid bilayer. This binding behaviour causes a decrease of the negative charge of the bilayer, as observed through ζ potential measurements, an increment in the mechanical resistance of the bilayer, as revealed by measurements of the breakthrough force applied with AFM and ζ potential measurements at high temperature, and a rearrangement of the spatial distances between ganglioside and cholesterol molecules in the LUVs, as determined by FRET measurements. These physicochemical changes are all known to impair the interaction of misfolded oligomers with cell membranes, protecting them from their toxicity. Taken together, our results illustrate how the incorporation in cell membranes of sterol molecules modified by the addition of polyamine tails leads to the modulation of physicochemical properties of the cell membranes themselves, making them more resistant to protein aggregates associated with neurodegeneration. More generally, they suggest that therapeutic strategies can be developed to reinforce cell membranes against protein misfolded assemblies.


 Please wait while we load your content...
Please wait while we load your content...