Predicting coated-nanoparticle drug release systems with perturbation-theory machine learning (PTML) models†
Abstract
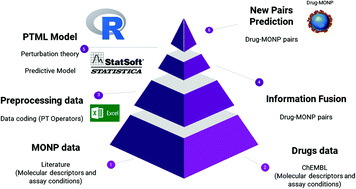
Nanoparticles (NPs) decorated with coating agents (polymers, gels, proteins, etc.) form Nanoparticle Drug Delivery Systems (DDNS), which are of high interest in nanotechnology and biomaterials science. There have been increasing reports of experimental data sets of biological activity, toxicity, and delivery properties of DDNS. However, these data sets are still dispersed and not as large as the datasets of DDNS components (NP and drugs). This has prompted researchers to train Machine Learning (ML) algorithms that are able to design new DDNS based on the properties of their components. However, most ML models reported up to date predictions of the specific activities of NP or drugs over a determined target or cell line. In this paper, we combine Perturbation Theory and Machine Learning (PTML algorithm) to train a model that is able to predict the best components (NP, coating agent, and drug) for DDNS design. In so doing, we downloaded a dataset of >30 000 preclinical assays of drugs from ChEMBL. We also downloaded an NP data set formed by preclinical assays of coated Metal Oxide Nanoparticles (MONPs) from public sources. Both the drugs and NP datasets of preclinical assays cover multiple conditions of assays that can be listed as two arrays, namely, cjdrug and cjNP. The cjdrug array includes >504 biological activity parameters (c0drug), >340 target proteins (c1drug), >650 types of cells (c2drug), >120 assay organisms (c3drug), and >60 assay strains (c4drug). On the other hand, the cjNP array includes 3 biological activity parameters (c0NP), 40 types of proteins (c1NP), 10 shapes of nanoparticles (c2NP), 6 assay media (c3NP), and 12 coating agents (c4NP). After downloading, we pre-processed both the data sets by separate calculation PT operators that are able to account for changes (perturbations) in the drug, coating agents, and NP chemical structure and/or physicochemical properties as well as for the assay conditions. Next, we carry out an information fusion process to form a final dataset of above 500 000 DDNS (drug + MONP pairs). We also trained other linear and non-linear PTML models using R studio scripts for comparative purposes. To the best of our knowledge, this is the first multi-label PTML model that is useful for the selection of drugs, coating agents, and metal or metal–oxide nanoparticles to be assembled in order to design new DDNS with optimal activity/toxicity profiles.


 Please wait while we load your content...
Please wait while we load your content...