3D domain swapping of azurin from Alcaligenes xylosoxidans†
Abstract
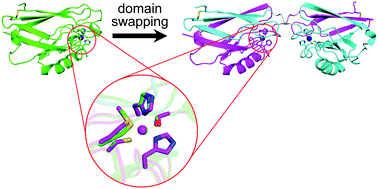
Protein oligomers have gained interest, owing to their increased knowledge in cells and promising utilization for future materials. Various proteins have been shown to 3D domain swap, but there has been no domain swapping report on a blue copper protein. Here, we found that azurin from Alcaligenes xylosoxidans oligomerizes by the procedure of 2,2,2-trifluoroethanol addition to Cu(I)-azurin at pH 5.0, lyophilization, and dissolution at pH 7.0, whereas it slightly oligomerizes when using Cu(II)-azurin. The amount of high order oligomers increased with the addition of Cu(II) ions to the dissolution process of a similar procedure for apoazurin, indicating that Cu(II) ions enhance azurin oligomerization. The ratio of the absorbance at 460 nm to that at ∼620 nm of the azurin dimer (Abs460/Abs618 = 0.113) was higher than that of the monomer (Abs460/Abs622 = 0.067) and the EPR A‖ value of the dimer (5.85 mT) was slightly smaller than that of the monomer (5.95 mT), indicating a slightly more rhombic copper coordination for the dimer. The redox potential of the azurin dimer was 342 ± 5 mV vs. NHE, which was 50 mV higher than that of the monomer. According to X-ray crystal analysis, the azurin dimer exhibited a domain-swapped structure, where the N-terminal region containing three β-strands was exchanged between protomers. The copper coordination structure was tetrahedrally distorted in the azurin dimer, similar to that in the monomer; however, the Cu–O(Gly45) bond length was longer for the dimer (monomer, 2.46–2.59 Å; dimer, 2.98–3.25 Å). These results open the door for designing oligomers of blue copper proteins by domain swapping.


 Please wait while we load your content...
Please wait while we load your content...