ProteoViz: a tool for the analysis and interactive visualization of phosphoproteomics data†
Abstract
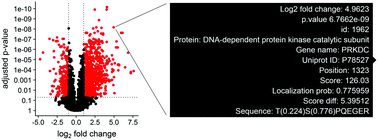
Quantitative proteomics generates large datasets with increasing depth and quantitative information. With the advance of mass spectrometry and increasingly larger data sets, streamlined methodologies and tools for analysis and visualization of phosphoproteomics are needed both at the protein and modified peptide levels. To assist in addressing this need, we developed ProteoViz, which includes a set of R scripts that perform normalization and differential expression analysis of both the proteins and enriched phosphorylated peptides, and identify sequence motifs, kinases, and gene set enrichment pathways. The tool generates interactive visualization plots that allow users to interact with the phosphoproteomics results and quickly identify proteins and phosphorylated peptides of interest for their biological study. The tool also links significant phosphosites with sequence motifs and pathways that will help explain the experimental conditions and guide future experiments. Here, we present the workflow and demonstrate its functionality by analyzing a phosphoproteomic data set from two lymphoma cell lines treated with kinase inhibitors. The scripts and data are freely available at https://github.com/ByrumLab/ProteoViz and via the ProteomeXchange with identifier PXD015606.


 Please wait while we load your content...
Please wait while we load your content...