Unraveling the RNA modification code with mass spectrometry
Abstract
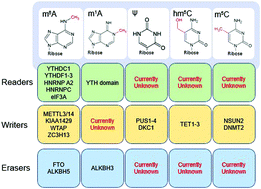
The discovery and analysis of modifications on proteins and nucleic acids has provided functional information that has rapidly accelerated the field of epigenetics. While protein post-translational modifications (PTMs), especially on histones, have been highlighted as critical components of epigenetics, the post-transcriptional modification of RNA has been a subject of more recently emergent interest. Multiple RNA modifications have been known to be present in tRNA and rRNA since the 1960s, but the exploration of mRNA, small RNA, and inducible tRNA modifications remains nascent. Sequencing-based methods have been essential to the field by creating the first epitranscriptome maps of m6A, m5C, hm5C, pseudouridine, and inosine; however, these methods possess significant limitations. Here, we discuss the past, present, and future of the application of mass spectrometry (MS) to the study of RNA modifications.


 Please wait while we load your content...
Please wait while we load your content...