Characterizing the length-dependence of DNA nanotube end-to-end joining rates†
Abstract
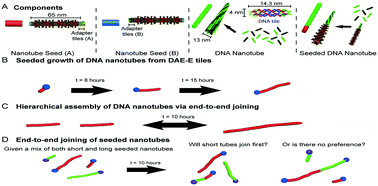
DNA nanotechnology offers a route towards the synthesis of custom nano-structured materials and circuits through hierarchical assembly processes. While predictive kinetic models are being developed for the assembly of DNA nanostructures from small monomeric components, a general model for the hierarchical assembly of DNA nanostructures remains elusive. DNA tile nanotubes provide an ideal model system for the study of hierarchical assembly via end-to-end joining. In this study, we experimentally characterize the length-dependence of the end-to-end joining rate of DNA tile nanotubes. We then test the ability of three different models of polymer end-to-end joining to reproduce experimentally measured changes in nanotube lengths during a joining reaction using an ODE model for nanotube joining. All three models predict physically realistic joining rates that are consistent with prior measurements, with a length-independent end-to-end joining rate model providing the best fit to the experimental data. A length-independent constant joining rate is consistent with other DNA self-assembly processes across a broad range of length scales and also suggests how tractable models for hierarchical DNA nanostructure could be developed.


 Please wait while we load your content...
Please wait while we load your content...