Single-cell microfluidics facilitates the rapid quantification of antibiotic accumulation in Gram-negative bacteria†
Abstract
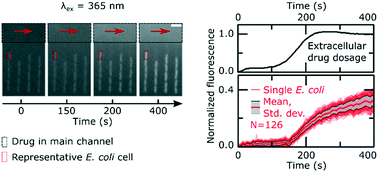
The double-membrane cell envelope of Gram-negative bacteria is a formidable barrier to intracellular antibiotic accumulation. A quantitative understanding of antibiotic transport in these cells is crucial for drug development, but this has proved elusive due to a dearth of suitable investigative techniques. Here we combine microfluidics and time-lapse auto-fluorescence microscopy to rapidly quantify antibiotic accumulation in hundreds of individual Escherichia coli cells. By serially manipulating the microfluidic environment, we demonstrated that stationary phase Escherichia coli, traditionally more refractory to antibiotics than growing cells, display reduced accumulation of the antibiotic ofloxacin compared to actively growing cells. Our novel microfluidic method facilitates the quantitative comparison of the role of the microenvironment versus that of the absence of key membrane transport pathways in cellular drug accumulation. Unlike traditional techniques, our assay is rapid, studying accumulation as the cells are dosed with the drug. This platform provides a powerful new tool for studying antibiotic accumulation in bacteria, which will be critical for the rational development of the next generation of antibiotics.


 Please wait while we load your content...
Please wait while we load your content...