Applications of the microphysiology systems database for experimental ADME-Tox and disease models†
Abstract
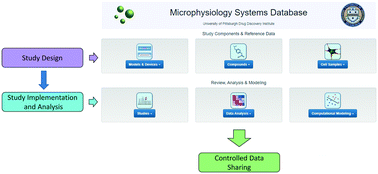
To accelerate the development and application of Microphysiological Systems (MPS) in biomedical research and drug discovery/development, a centralized resource is required to provide the detailed design, application, and performance data that enables industry and research scientists to select, optimize, and/or develop new MPS solutions, as well as to harness data from MPS models. We have previously implemented an open source Microphysiology Systems Database (MPS-Db), with a simple icon driven interface, as a resource for MPS researchers and drug discovery/development scientists (https://mps.csb.pitt.edu). The MPS-Db captures and aggregates data from MPS, ranging from static microplate models to integrated, multi-organ microfluidic models, and associates those data with reference data from chemical, biochemical, pre-clinical, clinical and post-marketing sources to support the design, development, validation, application and interpretation of the models. The MPS-Db enables users to manage their multifactor, multichip studies, then upload, analyze, review, computationally model and share data. Here we discuss how the sharing of MPS study data in the MS-Db is under user control and can be kept private to the individual user, shared with a select group of collaborators, or be made accessible to the general scientific community. We also present a test case using our liver acinus MPS model (LAMPS) as an example and discuss the use of the MPS-Db in managing, designing, and analyzing MPS study data, assessing the reproducibility of MPS models, and evaluating the concordance of MPS model results with clinical findings. We introduce the Disease Portal module with links to resources for the design of MPS disease models and studies and discuss the integration of computational models for the prediction of PK/PD and disease pathways using data generated from MPS models.
- This article is part of the themed collection: organ-on-a-chip systems: translating concept into practice


 Please wait while we load your content...
Please wait while we load your content...