Are 2D fingerprints still valuable for drug discovery?†
Abstract
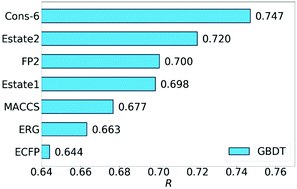
Recently, molecular fingerprints extracted from three-dimensional (3D) structures using advanced mathematics, such as algebraic topology, differential geometry, and graph theory have been paired with efficient machine learning, especially deep learning algorithms to outperform other methods in drug discovery applications and competitions. This raises the question of whether classical 2D fingerprints are still valuable in computer-aided drug discovery. This work considers 23 datasets associated with four typical problems, namely protein–ligand binding, toxicity, solubility and partition coefficient to assess the performance of eight 2D fingerprints. Advanced machine learning algorithms including random forest, gradient boosted decision tree, single-task deep neural network and multitask deep neural network are employed to construct efficient 2D-fingerprint based models. Additionally, appropriate consensus models are built to further enhance the performance of 2D-fingerprint-based methods. It is demonstrated that 2D-fingerprint-based models perform as well as the state-of-the-art 3D structure-based models for the predictions of toxicity, solubility, partition coefficient and protein–ligand binding affinity based on only ligand information. However, 3D structure-based models outperform 2D fingerprint-based methods in complex-based protein–ligand binding affinity predictions.
- This article is part of the themed collections: Emerging AI Approaches in Physical Chemistry, 2020 PCCP HOT Articles and PCCP Editor’s Choice, 2020


 Please wait while we load your content...
Please wait while we load your content...