Rosetta custom score functions accurately predict ΔΔG of mutations at protein–protein interfaces using machine learning†
Abstract
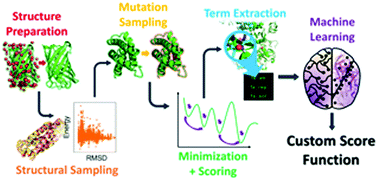
Protein–protein interfaces play essential roles in a variety of biological processes and many therapeutic molecules are targeted at these interfaces. However, accurate predictions of the effects of interfacial mutations to identify “hotspots” have remained elusive despite the myriad of modeling and machine learning methods tested. Here, for the first time, we demonstrate that nonlinear reweighting of energy terms from Rosetta, through the use of machine learning, exhibits improved predictability of ΔΔG values associated with interfacial mutations.


 Please wait while we load your content...
Please wait while we load your content...