Multiplex real-time PCR using double-strand primers and probes for the detection of nucleic acids†
Abstract
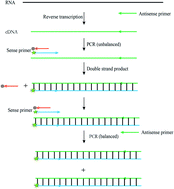
Multiplex PCR encounters difficulties in primer designing with all the primer pairs working at the same annealing temperature. In this study, we have developed a double-strand primer-mediated multiple strand displacement reaction for the detection of SARS-COV-2 ORF, N and E genes (as examples). The double primer is composed of a 5′-modified fluorophore strand, which does not impact polymerase extension and a 3′-modified quencher strand, which cannot impact elongation. At the annealing temperature, the fluorophore strand combined with the template, extended and resulted in fluorescence signal release. Results showed that the double-strand primer relatively exhibits a wide annealing temperature range and good compatibility between three pairs of primers and probes. These merits allow the simple and multiplex real-time fluorescence quantification of nucleic acids. The detection limit was 400 copies/mL, and the detection time was approximately 2 h. In addition to its extreme specificity and simplicity, this method has a wide range of applications such as multiple PCR and SNP detection.


 Please wait while we load your content...
Please wait while we load your content...