An approach for feature selection with data modelling in LC-MS metabolomics†
Abstract
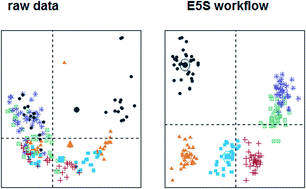
The data processing workflow for LC-MS based metabolomics study is suggested with signal drift correction, univariate analysis, supervised learning, feature selection and unsupervised modelling. The proposed approach requires only an annotation-free peak table and produces an extremely reduced set of the most relevant features together with validation via Receiver Operating Characteristic analysis for selected predictors, cross-validation and unsupervised projection. The presented study was initially optimised by its own experimental set and then was successfully tested by using 36 datasets from 21 publicly available metabolomics projects. The suggested workflow can be used for classification purposes in high dimensional metabolomics studies and as a first step in exploratory analysis, data projection, biomarker selection, data integration and fusion.


 Please wait while we load your content...
Please wait while we load your content...