Development of a non-targeted high-coverage microbial metabolomics pretreatment method and its application to drug resistant Salmonella†
Abstract
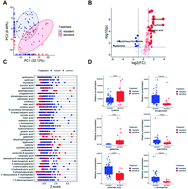
Metabolomics as an effective omics research tool is widely used in the study of pathogenic bacteria drug resistance mechanisms. The pretreatment methods affect the coverage and abundance of metabolites, leading to a diversity of results. Therefore, it is necessary to develop a high-coverage and robust pretreatment method to analyze and study resistant-related metabolites (carbohydrates, lipids, and amino acids). The four essential pretreatment steps of Salmonella for the GC/MS-based metabolomics study were optimized, including quenching (methanol–water and liquid nitrogen), solvent extraction (acetonitrile–isopropanol–water, methyl tert-butyl ether (MTBE) and methanol–chloroform), cell disruption (stainless-steel ball grinding and liquid nitrogen milling) and derivatization (normal-temperature and high-temperature derivatization). Metabolite profiling was performed to investigate metabolic differences between different methods. The optimized pretreatment method for the non-targeted metabolic profiling of drug-resistant Salmonella was as follows: liquid-nitrogen quenching, acetonitrile–isopropanol–water (3 : 3 : 2, v/v) as an extracting agent, stainless-steel ball grinding for cell disruption and derivatization at 37 °C. The optimized pretreatment method will provide more types of metabolites, a higher abundance of metabolites, and better repeatability and stability. The optimized method was implemented for the metabolic profiling of the resistance and sensitivity of Salmonella with serotype Derby. The method has the potential to be applied to the study of the metabolism of other pathogenic bacteria.


 Please wait while we load your content...
Please wait while we load your content...