Centrifugal microfluidic lab-on-a-chip system with automated sample lysis, DNA amplification and microarray hybridization for identification of enterohemorrhagic Escherichia coli culture isolates†
Abstract
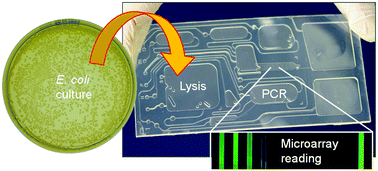
The development of technology for the rapid, automated identification of bacterial culture isolates can help regulatory agencies to shorten response times in food safety surveillance, compliance, and enforcement as well as outbreak investigations. While molecular methods such as polymerase chain reaction (PCR) enable the identification of microbial organisms with high sensitivity and specificity, they generally rely on sophisticated instrumentation and elaborate workflows for sample preparation with an undesirably high level of hands-on engagement. Herein, we describe the design, operation and performance of a lab-on-a-chip system integrating thermal lysis, PCR amplification and microarray hybridization on the same cartridge. The assay is performed on a centrifugal microfluidic platform that allows for pneumatic actuation of liquids during rotation, making it possible to perform all fluidic operations in a fully-automated fashion without the need for integrating active control elements on the microfluidic cartridge. The cartridge, which is fabricated from hard and soft thermoplastic polymers, is compatible with high-volume manufacturing (e.g., injection molding). Chip design and thermal interface were both optimized to ensure efficient heat transfer and allow for fast thermal cycling during the PCR process. The integrated workflow comprises 14 steps and takes less than 2 h to complete. The only manual steps are related to loading of the sample and reagents on the cartridge as well as fluorescence imaging of the microarray. On-chip lysis and PCR amplification both provided results comparable to those obtained by bench-top instrumentation. The microarray, incorporating a panel of oligonucleotide probes for multiplexed detection of seven enterohemorrhagic E. coli priority serotypes, was implemented on a cyclic olefin copolymer substrate using a novel activation scheme that involves the conversion of hydroxyl groups (derived from oxygen plasma treatment) into reactive cyanate ester using cyanogen bromide. On-chip hybridization was demonstrated in a non-quantitative fashion using fluorescently-labelled gene markers for E. coli O157:H7 (rfbO157, eae, vt1, and vt2) obtained through a multiplexed PCR amplification step.


 Please wait while we load your content...
Please wait while we load your content...