Deep learning networks for the recognition and quantitation of surface-enhanced Raman spectroscopy†
Abstract
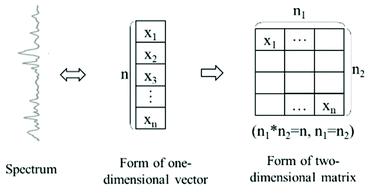
Surface-enhanced Raman spectroscopy (SERS) based on machine learning methods has been applied in material analysis, biological detection, food safety, and intelligent analysis. However, machine learning methods generally require extra preprocessing or feature engineering, and handling large-scale data using these methods is challenging. In this study, deep learning networks were used as fully connected networks, convolutional neural networks (CNN), fully convolutional networks (FCN), and principal component analysis networks (PCANet) to determine their abilities to recognise drugs in human urine and measure pirimiphos-methyl in wheat extract in the two input forms of a one-dimensional vector or a two-dimensional matrix. The best recognition result for drugs in urine with an accuracy of 98.05% in the prediction set was obtained using CNN with spectra as input in the matrix form. The optimal quantitation for pirimiphos-methyl was obtained using FCN with spectra in the matrix form, and the analysis was accomplished with a determination coefficient of 0.9997 and a root mean square error of 0.1574 in the prediction set. These networks performed better than the common machine learning methods. Overall, the deep learning networks provide feasible alternatives for the recognition and quantitation of SERS.


 Please wait while we load your content...
Please wait while we load your content...