Systematic bioinformatic analysis of nutrigenomic data of flavanols in cell models of cardiometabolic disease†
Abstract
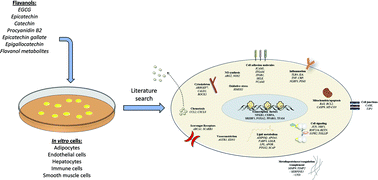
Flavanol intake positively influences several cardiometabolic risk factors in humans. However, the specific molecular mechanisms of action of flavanols, in terms of gene regulation, in the cell types relevant to cardiometabolic disease have never been systematically addressed. On this basis, we conducted a systematic literature review and a comprehensive bioinformatic analysis of genes whose expression is affected by flavanols in cells defining cardiometabolic health: hepatocytes, adipocytes, endothelial cells, smooth muscle cells and immune cells. A systematic literature search was performed using the following pre-defined criteria: treatment with pure compounds and metabolites (no extracts) at low concentrations that are close to their plasma concentrations. Differentially expressed genes were analyzed using bioinformatics tools to identify gene ontologies, networks, cellular pathways and interactions, as well as transcriptional and post-transcriptional regulators. The systematic literature search identified 54 differentially expressed genes at the mRNA level in in vitro models of cardiometabolic disease exposed to flavanols and their metabolites. Global bioinformatic analysis revealed that these genes are predominantly involved in inflammation, leukocyte adhesion and transendothelial migration, and lipid metabolism. We observed that, although the investigated cells responded differentially to flavanol exposure, the involvement of anti-inflammatory responses is a common mechanism of flavanol action. We also identified potential transcriptional regulators of gene expression: transcriptional factors, such as GATA2, NFKB1, FOXC1 or PPARG, and post-transcriptional regulators: miRNAs, such as mir-335-5p, let-7b-5p, mir-26b-5p or mir-16-5p. In parallel, we analyzed the nutrigenomic effects of flavanols in intestinal cells and demonstrated their predominant involvement in the metabolism of circulating lipoproteins. In conclusion, the results of this systematic analysis of the nutrigenomic effects of flavanols provide a more comprehensive picture of their molecular mechanisms of action and will support the future setup of genetic studies to pave the way for individualized dietary recommendations.
- This article is part of the themed collections: Food & Function Recent HOT articles and International Conference on Polyphenols and Health (ICPH2019) collection


 Please wait while we load your content...
Please wait while we load your content...