Insights into origins and function of the unexplored majority of the reductive dehalogenase gene family as a result of genome assembly and ortholog group classification†
Abstract
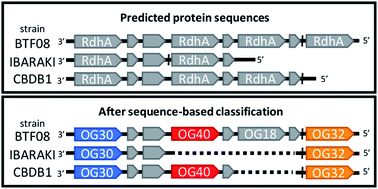
Organohalide respiring bacteria (OHRB) express reductive dehalogenases for energy conservation and growth. Some of these enzymes catalyze the reductive dehalogenation of chlorinated and brominated pollutants in anaerobic subsurface environments, providing a valuable ecosystem service. Dehalococcoides mccartyi strains have been most extensively studied owing to their ability to dechlorinate all chlorinated ethenes – most notably carcinogenic vinyl chloride – to ethene. The genomes of OHRB, particularly obligate OHRB, often harbour multiple putative reductive dehalogenase genes (rdhA), most of which have yet to be characterized. We recently sequenced and closed the genomes of eight new strains, increasing the number of available D. mccartyi genomes in NCBI from 16 to 24. From all available OHRB genomes, we classified predicted translations of reductive dehalogenase genes using a previously established 90% amino acid pairwise identity cut-off to identify Ortholog Groups (OGs). Interestingly, the majority of D. mccartyi dehalogenase gene sequences, once classified into OGs, exhibited a remarkable degree of synteny (gene order) in all genomes sequenced to date. This organization was not apparent without the classification. A high degree of synteny indicates that differences arose from rdhA gene loss rather than recombination. Phylogenetic analysis suggests that most rdhA genes have a long evolutionary history in the Dehalococcoidia with origin prior to speciation of Dehalococcoides and Dehalogenimonas. We also looked for evidence of synteny in the genomes of other species of OHRB. Unfortunately, there are too few closed Dehalogenimonas genomes to compare at this time. There is some partial evidence for synteny in the Dehalobacter restrictus genomes, but here too more closed genomes are needed for confirmation. Interestingly, we found that the rdhA genes that encode enzymes that catalyze dehalogenation of industrial pollutants are the only rdhA genes with strong evidence of recent lateral transfer – at least in the genomes examined herein. Given the utility of the RdhA sequence classification to comparative analyses, we are building a public web server (http://RDaseDB.biozone.utoronto.ca) for the community to use, which allows users to add and classify new sequences, and download the entire curated database of reductive dehalogenases.
- This article is part of the themed collection: Halogenated (semi)volatile organic compounds (“X(S)VOCs”)


 Please wait while we load your content...
Please wait while we load your content...