Dynamical properties of enzyme–substrate complexes disclose substrate specificity of the SARS-CoV-2 main protease as characterized by the electron density descriptors†
Abstract
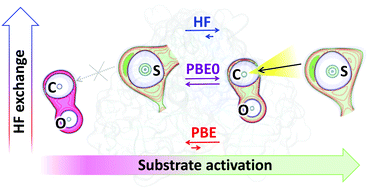
A dynamical approach is proposed to discriminate between reactive (rES) and nonreactive (nES) enzyme–substrate complexes taking the SARS-CoV-2 main protease (Mpro) as an important example. Molecular dynamics simulations with the quantum mechanics/molecular mechanics potentials (QM(DFT)/MM-MD) followed by the electron density analysis are employed to evaluate geometry and electronic properties of the enzyme with different substrates along MD trajectories. We demonstrate that mapping the Laplacian of the electron density and the electron localization function provides easily visible images of the substrate activation that allow one to distinguish rES and nES. The computed fractions of reactive enzyme–substrate complexes along MD trajectories well correlate with the findings of recent experimental studies on the substrate specificity of Mpro. The results of our simulations demonstrate the role of the theory level used in QM subsystems for a proper description of the nucleophilic attack of the catalytic cysteine residue in Mpro. The activation of the carbonyl group of a substrate is correctly characterized with the hybrid DFT functional PBE0, whereas the use of a GGA-type PBE functional, that lacks the admixture of the Hartree–Fock exchange fails to describe activation.
- This article is part of the themed collection: 2020 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...