Theoretical understanding of the thermodynamics and interactions in transcriptional regulator TtgR–ligand binding†
Abstract
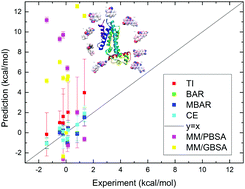
The transcriptional regulator TtgR belongs to the TetR family of transcriptional repressors. It depresses the transcription of the TtgABC operon and itself and thus regulates the extrusion of noxious chemicals with efflux pumps in bacterial cells. As the ligand-binding domain of TtgR is rather flexible, it can bind with a number of structurally diverse ligands, such as antibiotics, flavonoids and aromatic solvents. In the current work, we perform equilibrium and nonequilibrium alchemical free energy simulation to predict the binding affinities of a series of ligands targeting the TtgR protein and an agreement between the theoretical prediction and the experimental result is observed. End-point methods MM/PBSA and MM/GBSA are also employed for comparison. We further study the interaction maps and contacts between the protein and the ligand and identify important interactions in the protein–ligand binding cases. The dynamics fluctuation and secondary structures are also investigated. The current work sheds light on atomic and thermodynamic understanding of the TtgR–ligand interactions.


 Please wait while we load your content...
Please wait while we load your content...