Computational approaches to studying methylated H4K20 recognition by DNA repair factor 53BP1
Abstract
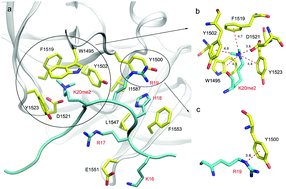
Histone lysine methylation regulates the recruitment of mammalian DNA repair factor 53BP1 to the histone H4 lysine 20 (H4K20), through specific recognition of the tandem Tudor domain of 53BP1. The di- and mono-methylated H4K20 bind to 53BP1 with high affinity, but the non- and tri-methylated H4K20 do not. Here, we develop a new approach to carry out computational study to unravel the binding mechanism of methylated H4K20 by 53BP1 and to compute relative binding affinities of different methylations of H4K20 by 53BP1. First, hot spots in 53BP1 were predicted by computational alanine scanning and aromatic cages formed by W1495, Y1500, Y1502, and Y1523 are found to provide the dominant binding to di- and mono-methylated H4K20 in addition to D1521. Secondly, a de-methylation method is proposed to predict relative binding free energies between 53BP1 and different methylated states of H4K20. Finally, the tri-methylated and non-methylated H4K20/53BP1 complexes are found to be dynamically unstable, explaining the experimental finding that neither can bind to 53BP1. The present work provides an important theoretical basis for our understanding of histone methylations of H4K20 and their recognition mechanism by DNA repair factor 53BP1.

 Please wait while we load your content...
Please wait while we load your content...