Comparative study of serum sample preparation methods in aggregation-based plasmonic sensing†
Abstract
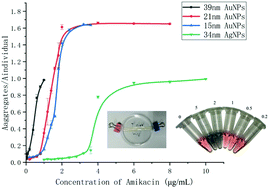
The use of nanoparticle-based colorimetric methods has received considerable attention in a broad range of clinical and biomedical applications due to their high sensitivity, low cost, extreme simplicity and excellent analytical performance. However, the formation of a protein corona has severely limited the application of nanoparticles (NPs) in clinical samples, which can confer colloidal stability to serum-exposed nanoparticles compared to pristine particles. To address this challenge, dialysis, ultrafiltration and phenol : chloroform : isopentanol extraction methods were compared aiming at facile and routine protein separation methods to eliminate the formation of protein corona on NPs and the development of a sensitive and simple therapeutic drug monitoring (TDM) assay for the detection of aminoglycoside antibiotics in serum. Based on the comparison of the sensitivity of the localized surface plasmon resonance (LSPR) aggregation assay in pure water, untreated serum and serum after the different sample preparation methods, we revealed by Coomassie blue staining that proteins in the serum were the predominant interfering molecules to degrade the sensitivity of serum-based aggregation assays. Using dialysis, naked eye semi-quantification was achieved at the clinical level for amikacin, tobramycin and streptomycin. The dialysis efficiency and dialysis coefficient of amikacin were also measured to prove the efficacy of dialysis as a fast and efficient protein-removal method. This strategy is expected to be applicable universally as a pretreatment for the assay of small molecules with plasmonic assays in crude biological samples.


 Please wait while we load your content...
Please wait while we load your content...