Substrate-rigidity dependent migration of an idealized twitching bacterium
Abstract
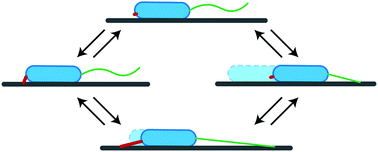
Mechanical properties of the extracellular matrix are important determinants of cellular migration in diverse processes, such as immune response, wound healing, and cancer metastasis. Moreover, recent studies indicate that even bacterial surface colonization can depend on the mechanics of the substrate. Here, we focus on physical mechanisms that can give rise to substrate-rigidity dependent migration. We study a “twitcher”, a cell driven by extension–retraction cycles, to idealize bacteria and perhaps eukaryotic cells that employ a slip-stick mode of motion. The twitcher is asymmetric and always pulls itself forward at its front. Analytical calculations show that the migration speed of a twitcher depends non-linearly on substrate rigidity. For soft substrates, deformations do not lead to build-up of significant force and the migration speed is therefore determined by stochastic adhesion unbinding. For rigid substrates, forced adhesion rupture determines the migration speed. Depending on the force-sensitivity of front and rear adhesions, forced bond rupture implies an increase or a decrease of the migration speed. A requirement for the occurrence of rigidity-dependent stick-slip migration is a “sticky” substrate, with binding rates being an order of magnitude larger than unbinding rates in absence of force. Computer simulations show that small stall forces of the driving machinery lead to a reduced movement on high rigidities, regardless of force-sensitivities of bonds. The simulations also confirm the occurrence of rigidity-dependent migration speed in a generic model for slip-stick migration of cells on a sticky substrate.


 Please wait while we load your content...
Please wait while we load your content...