In vivo methylation of OLA1 revealed by activity-based target profiling of NTMT1†
Abstract
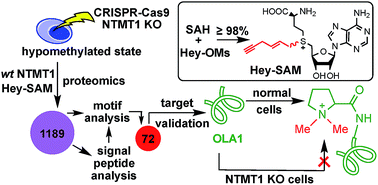
N-Terminal methyltransferase 1 (NTMT1) catalyzes the N-terminal methylation of proteins with a specific N-terminal motif after methionine removal. Aberrant N-terminal methylation has been implicated in several cancers and developmental diseases. Together with motif sequence and signal peptide analyses, activity-based substrate profiling of NTMT1 utilizing (E)-hex-2-en-5-ynyl-S-adenosyl-L-methionine (Hey-SAM) revealed 72 potential targets, which include several previously confirmed ones and many unknowns. Target validation using normal and NTMT1 knock-out (KO) HEK293FT cells generated by CRISPR-Cas9 demonstrated that Obg-like ATPase 1 (OLA1), a protein involved in many critical cellular functions, is methylated in vivo by NTMT1. Additionally, Hey-SAM synthesis achieved ≥98% yield for SAH conversion.


 Please wait while we load your content...
Please wait while we load your content...