Chemical labeling for fine mapping of IgG N-glycosylation by ETD-MS†
Abstract
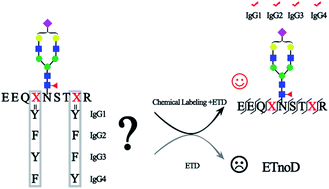
Immunoglobulin G (IgG), which contains four subclasses (IgG1–4), is one of the most important classes of glycoproteins in the immune system. Because of its importance in the immune system, a steady increase of interest in developing IgG as the biomarker or biotherapeutic agent for the treatment of diseases has been seen, as most therapeutic mAbs were IgG-based. N-Glycosylation of IgG is crucial for its effector function and makes IgG highly heterogeneous both in structure and function, although all four subclasses of IgG contain only a single N-glycosylation site in the Fc region with a highly similar amino acid sequence. Therefore, fine mapping of IgG glycosylation is necessary for understanding the IgG function and avoiding aberrant glycosylation in mAbs. However, site-specific and comprehensive N-glycosylation analysis of IgG subclasses still cannot be achieved by MS alone due to the partial sequence coverage and loss of connections among glycosylation of the protein sequence. We report here a chemical labeling strategy to improve the electron transfer dissociation efficiency in mass spectrometry analysis, which enables a 100% peptide sequence coverage of N-glycopeptides in all subclasses of IgG. Combined with high-energy collisional dissociation for the fragmentation of glycans, fine mapping of the N-glycosylation profile of IgG is achieved. This comprehensive glycosylation analysis strategy for the first time allows the discrimination of IgG3 and IgG4 intact N-glycopeptides with high similarity in sequence without the antibody-based pre-separation. Using this strategy, aberrant serum IgG N-glycosylation for four IgG subclasses associated with cirrhosis and hepatocellular carcinoma was revealed. Moreover, this method identifies 5 times more intact glycopeptides from human serum than the native-ETD method, implying that the approach can also accommodate large-scale site-specific profiling of glycoproteomes.


 Please wait while we load your content...
Please wait while we load your content...