PEGylated surfaces for the study of DNA–protein interactions by atomic force microscopy†
Abstract
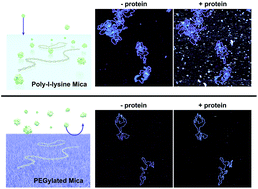
DNA–protein interactions are vital to cellular function, with key roles in the regulation of gene expression and genome maintenance. Atomic force microscopy (AFM) offers the ability to visualize DNA–protein interactions at nanometre resolution in near-physiological buffers, but it requires that the DNA be adhered to the surface of a solid substrate. This presents a problem when working in biologically relevant protein concentrations, where proteins may be present in large excess in solution; much of the biophysically relevant information can therefore be occluded by non-specific protein binding to the underlying substrate. Here we explore the use of PLLx-b-PEGy block copolymers to achieve selective adsorption of DNA on a mica surface for AFM studies. Through varying both the number of lysine and ethylene glycol residues in the block copolymers, we show selective adsorption of DNA on mica that is functionalized with a PLL10-b-PEG113/PLL1000–2000 mixture as viewed by AFM imaging in a solution containing high concentrations of streptavidin. We show – through the use of biotinylated DNA and streptavidin – that this selective adsorption extends to DNA–protein complexes and that DNA-bound streptavidin can be unambiguously distinguished in spite of an excess of unbound streptavidin in solution. Finally, we apply this to the nuclear enzyme PARP1, resolving the binding of individual PARP1 molecules to DNA by in-liquid AFM.
- This article is part of the themed collection: Editor’s Choice: Recent breakthroughs in nanobiotechnology


 Please wait while we load your content...
Please wait while we load your content...